SeeMost radiomic features depend on the data acquisition settings, including the scanner brand, the imaging and image reconstruction, and voxel size settings. It is therefore recommended to study radiomic features from images that have been acquired using the exact same scanner and protocol settings. If this is not possible, and if you want to combine radiomic features obtained using different settings in a single analysis, some feature harmonization should be performed.
For PET and CT data, we suggest to harmonize radiomic features using the ComBat approach, that we previously validated (http://jnm.snmjournals.org/content/59/8/1321.long, https://doi.org/10.1148/radiol.2019182023), and that is freely available as a R routine : https://github.com/Jfortin1/ComBatHarmonization
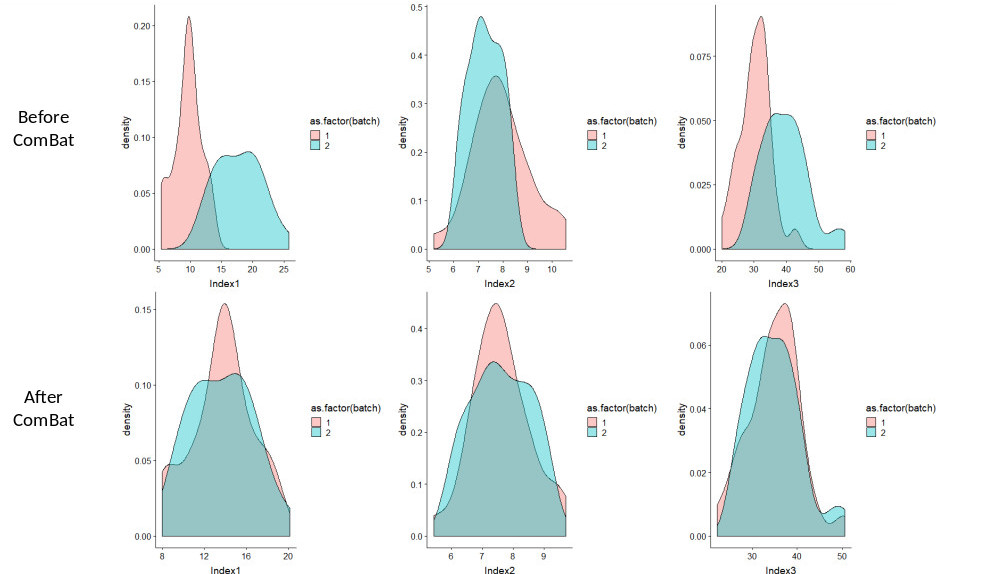
We provide below an example of R script and two sample files (before and after ComBat) involving 3 features, 30 regions of interest for scanner 1 and 30 regions of interest for scanner 2. The code corresponds to using ComBat without any covariate. Before ComBat, the statistical distributions of the 3 features are quite different between scanner 1 (pink) and scanner 2 (cyan) (top row), while after ComBat, the distributions from the two scanners better match for the 3 features.
CombaTool is available here: https://forlhac.shinyapps.io/Shiny_ComBat
A guide to ComBat harmonization of imaging biomarkers in multicenter studies is available here: https://jnm.snmjournals.org/content/63/2/172.abstract
