 |
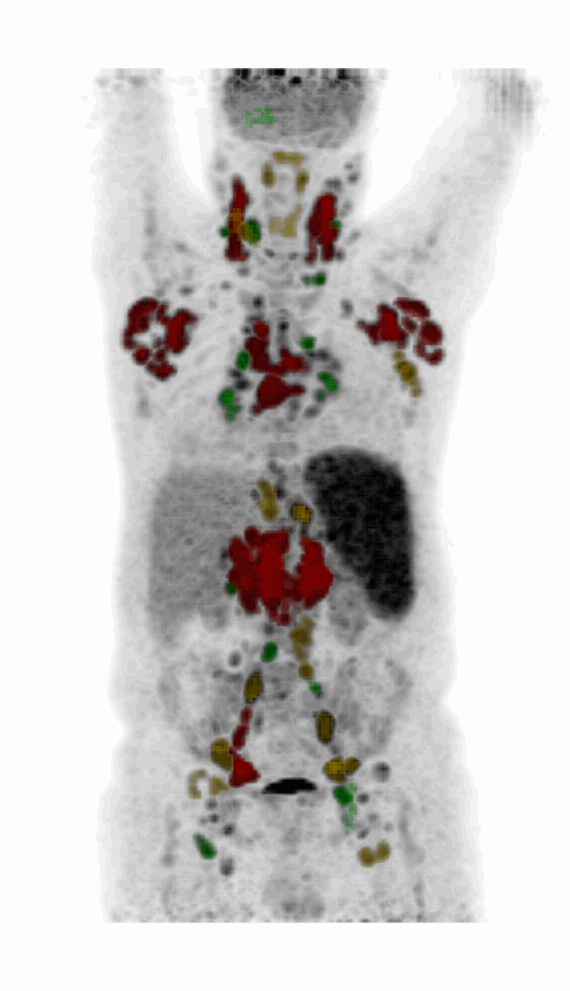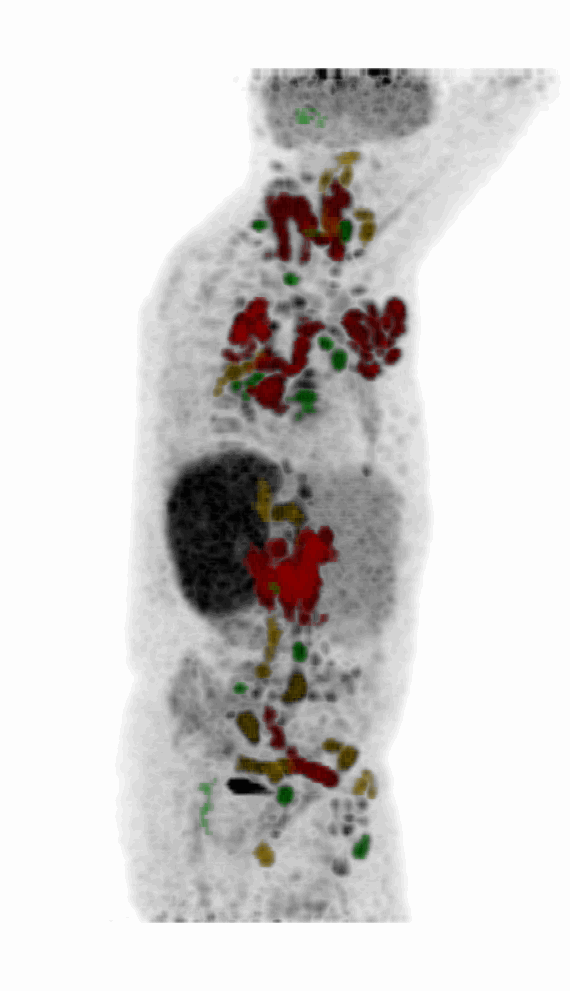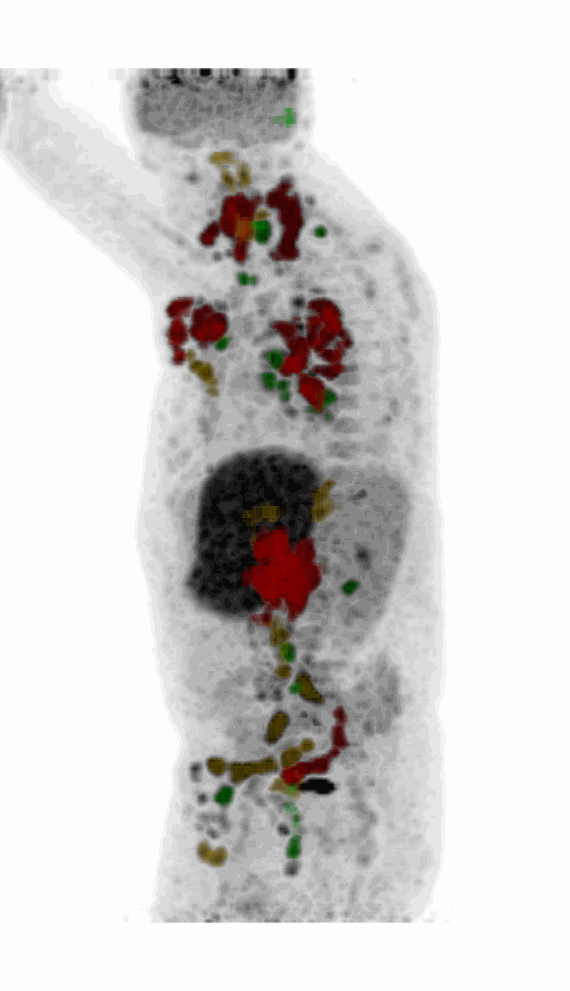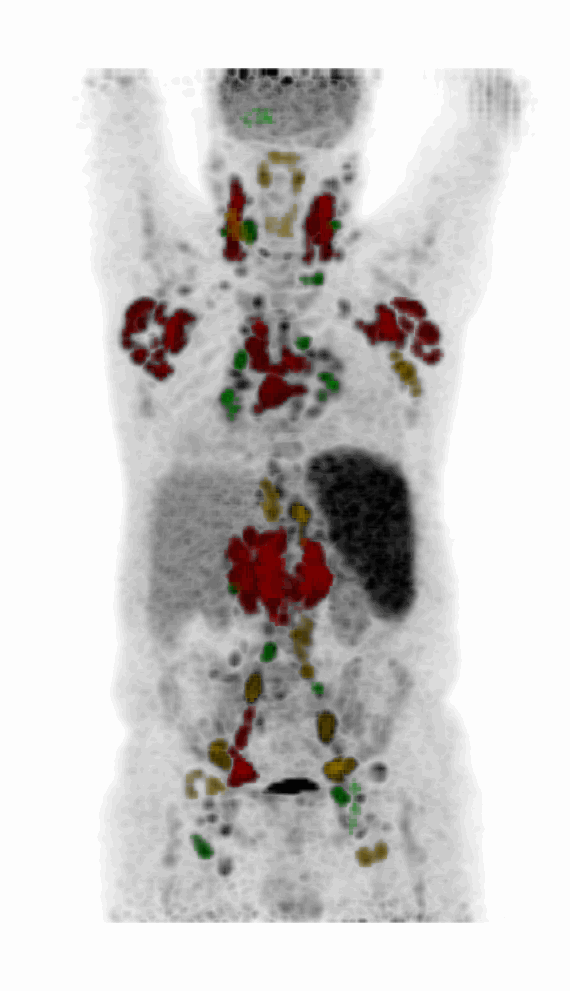 |
 |
| MIP PET/CT | MIP PET/ROI | MIP PET |
To view the LIFEx license: License
The libraries used by LIFEx and included in the package are as follows (you do NOT have anything to install except the single LIFEx package):


GENERAL TERMS OF LICENSE FOR LIFEx SOFTWARE
You belong to a private company: see this portal
Last update: June 15th, 2023
USE OF THE LIFEx SOFTWARE IS SUBJECT TO THE FOLLOWING TERMS AND CONDITIONS. IF YOU DO NOT ACCEPT THESE TERMS AND CONDITIONS, DO NOT DOWNLOAD THE SOFTWARE.
USERS ARE NOTIFIED THAT THE SOFTWARE AND THE RISKS INHERENT TO ITS USE, MODIFICATION AND/OR DEVELOPMENT AND REPRODUCTION ARE NOT COVERED BY ANY WARRANTY FROM LICENSOR. IT IS HIGHLY RECOMMENDED THAT YOU ONLY LOAD AND/OR USE THE SOFTWARE IF YOU HAVE SPECIALIZED IN-DEPTH INFORMATION TECHNOLOGY KNOW-HOW.
By downloading the Software, the Licensee accepts the terms of this License. If Licensee does not accept the License terms, then Licensee shall not use the Software.
The Licensee acknowledges that the License shall prevail on any conflicting terms provided by Licensee (such as general terms of purchase), unless otherwise specifically agreed in writing by Licensor.
Licensor reserves the right to modify or replace the License by different terms. However, the contractual terms applicable to Licensee will be the License in force at the time of Licensee’s last download of the Software.

Introduction
Texture analysis is gaining considerable interest in medical imaging, particularly for identifying parameters that characterize tumor heterogeneity. We developed easy-to-use freeware that enables the calculation of a wide range of conventional, texture, and shape indices from PET, MRI, ultrasound, and CT images.
The software is written in Java and does not rely on any commercial libraries. LIFEx is designed for researchers, radiologists, nuclear medicine physicians, and oncologists, providing access to 311 histogram, texture, and shape indices in addition to conventional metrics. Users can customise calculation settings, such as the resampling method and the number of gray levels used for texture analysis.