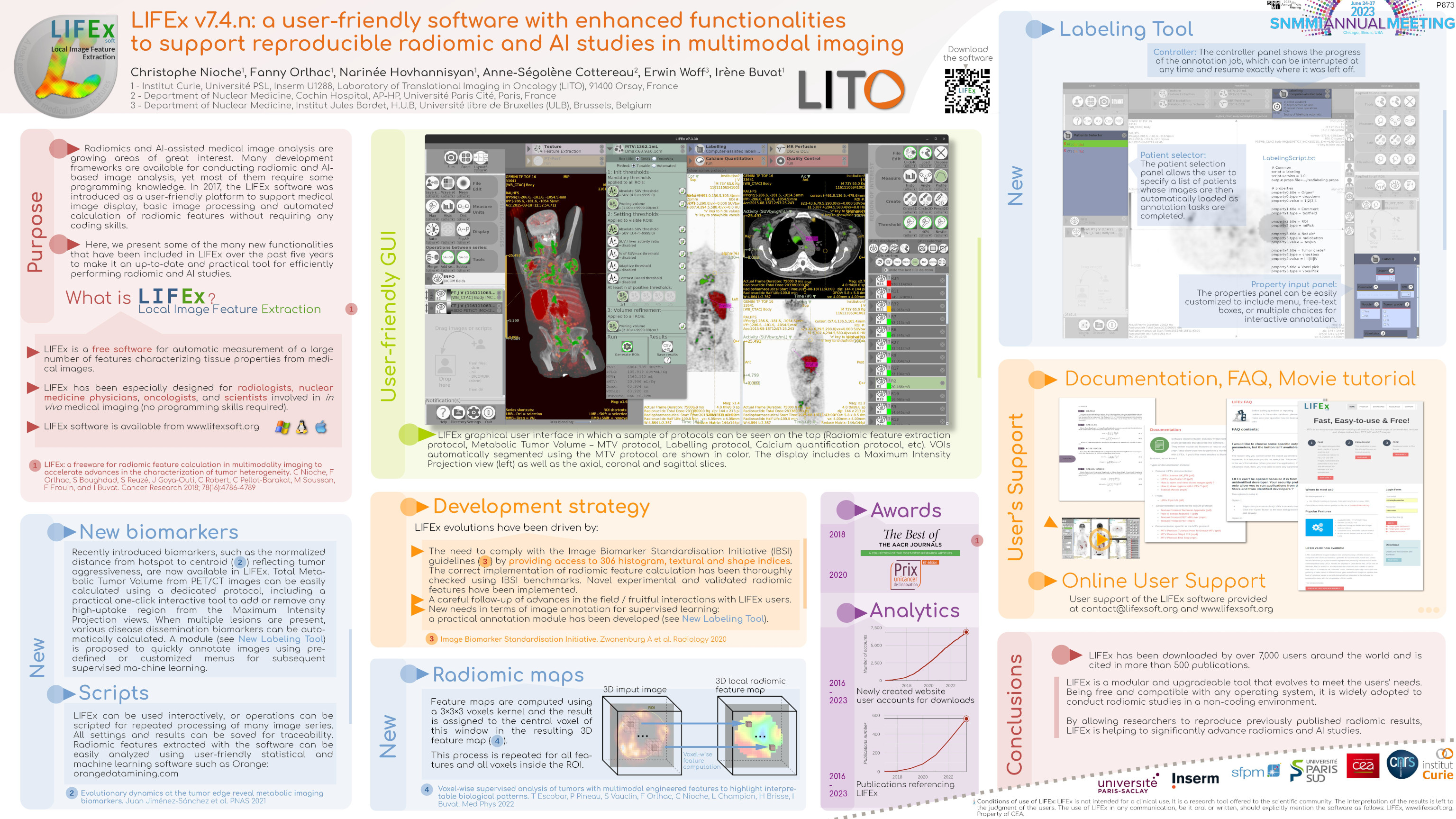
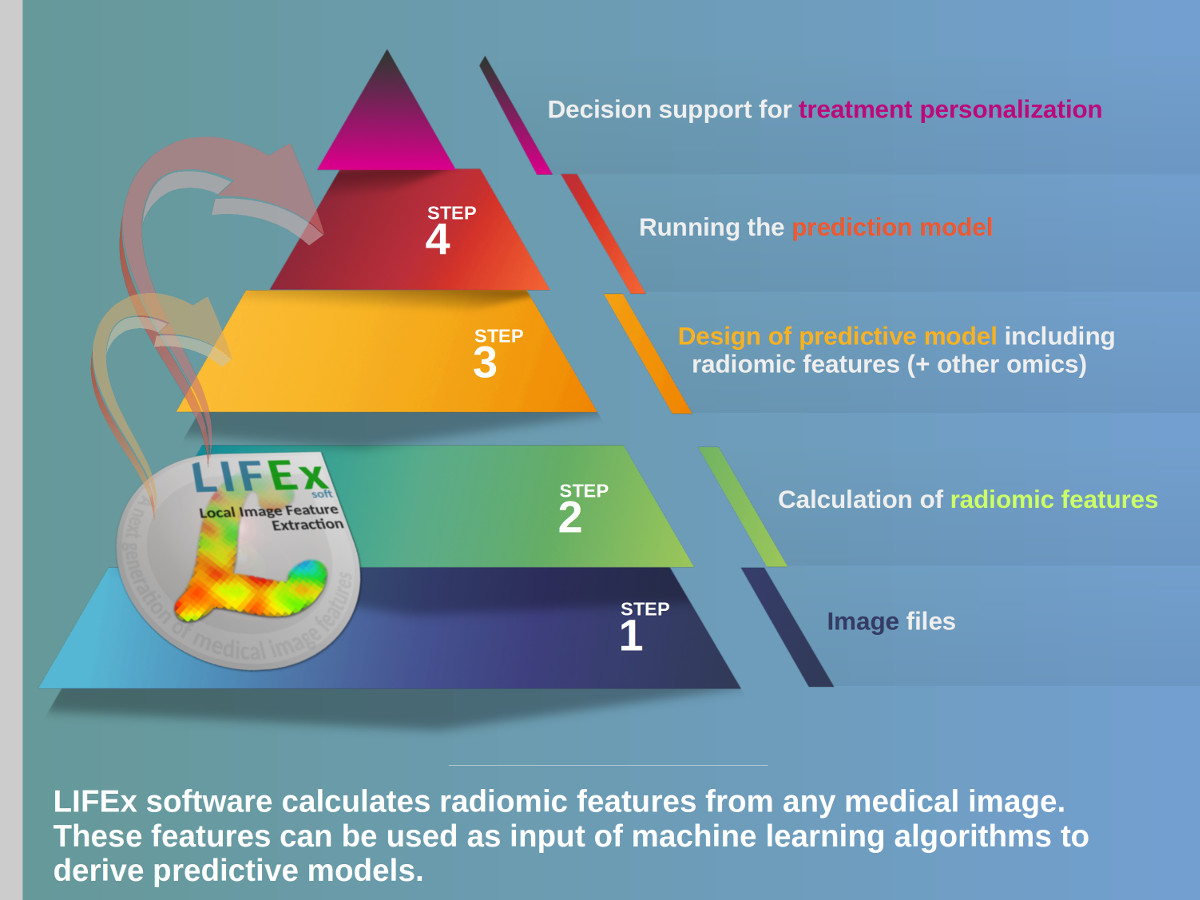
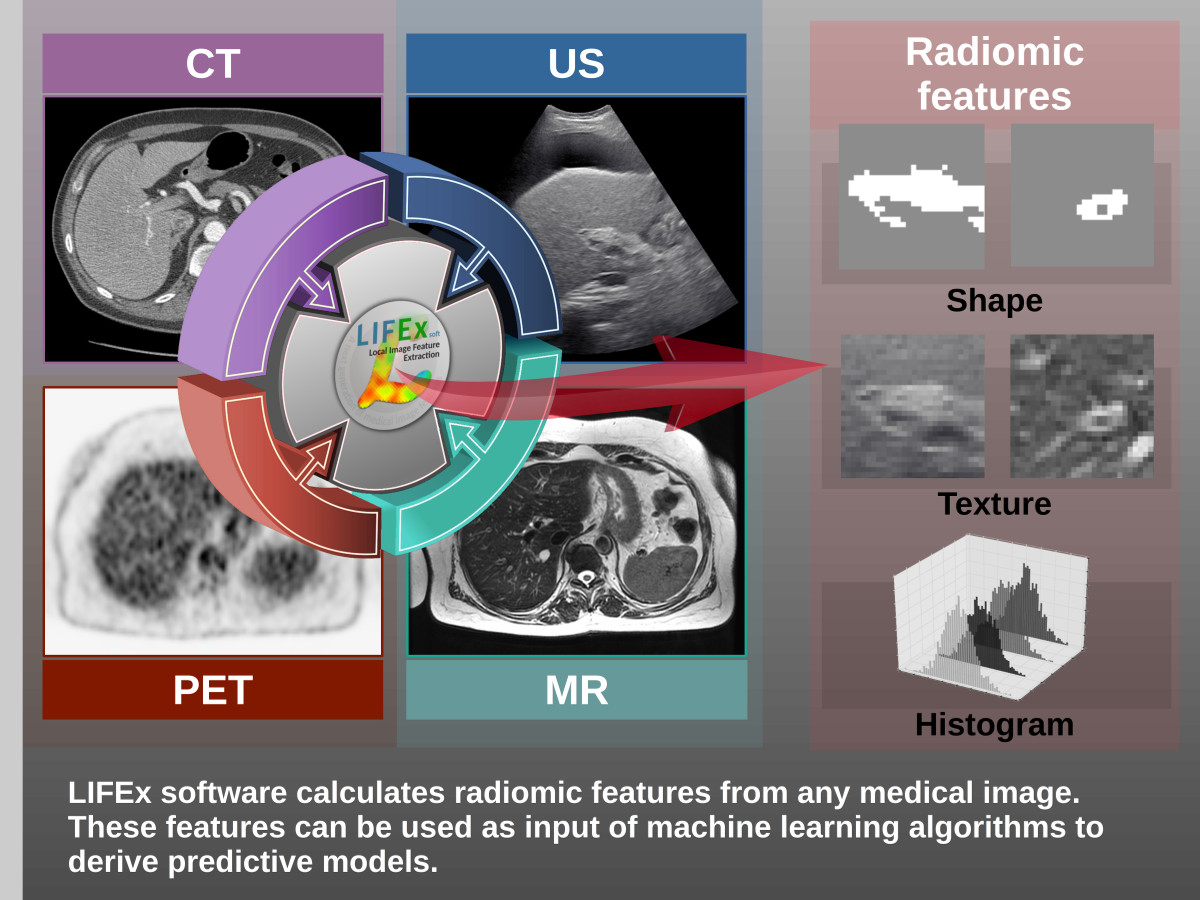
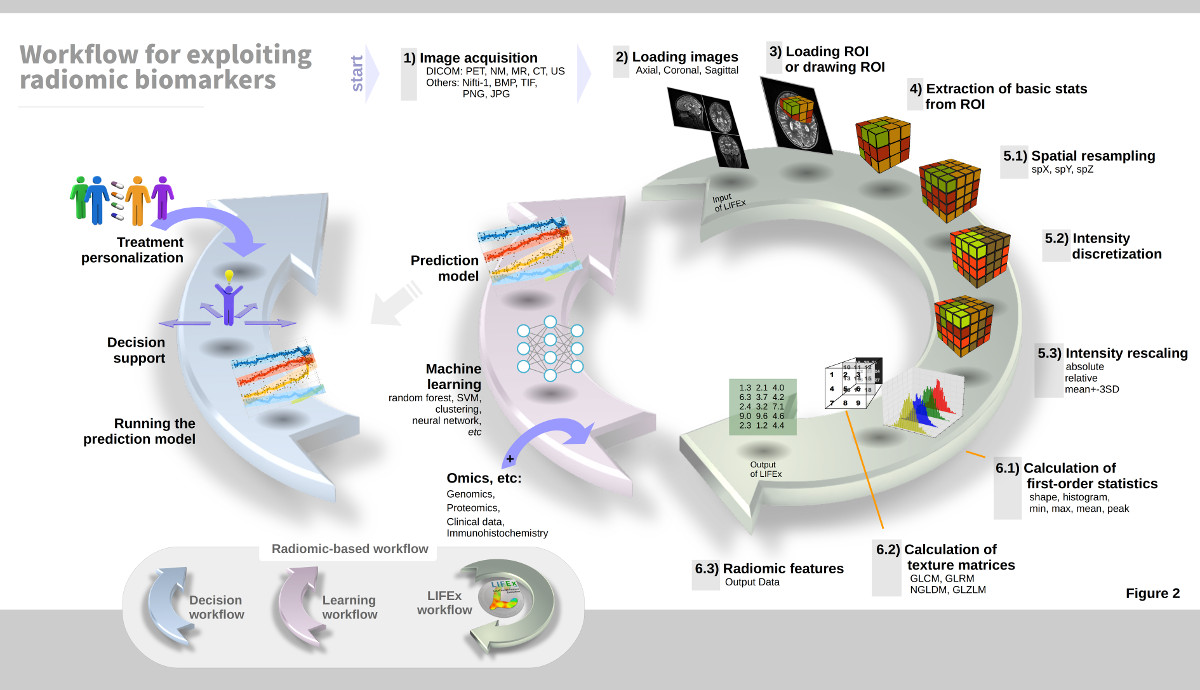
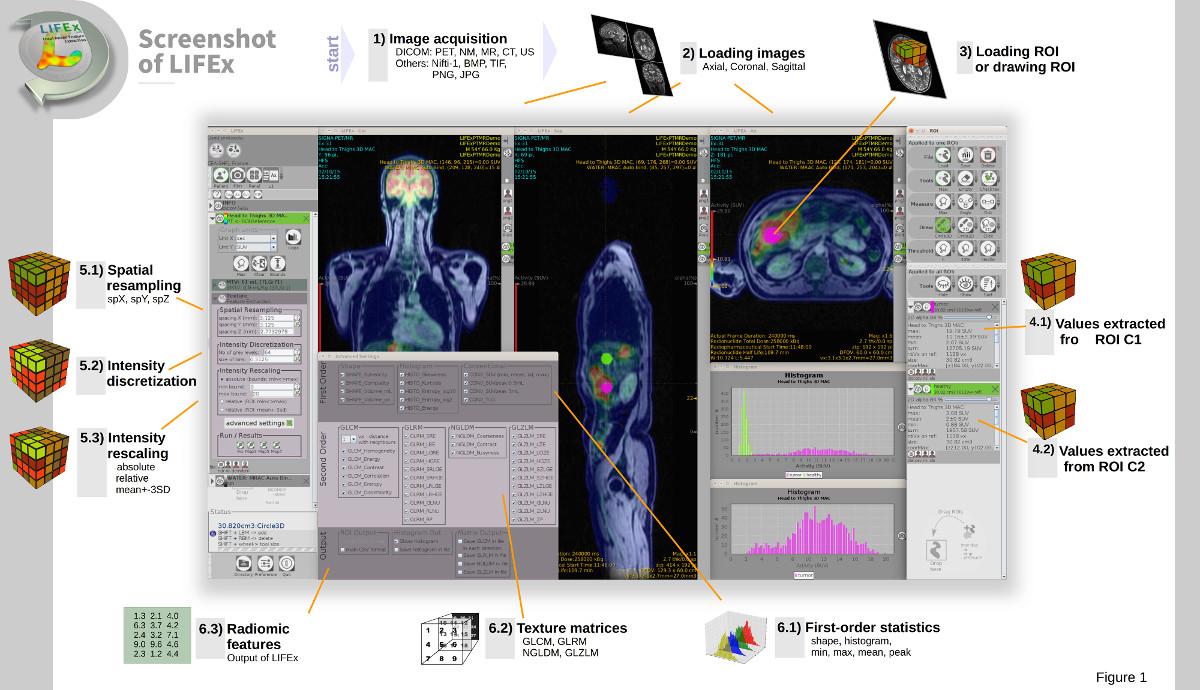
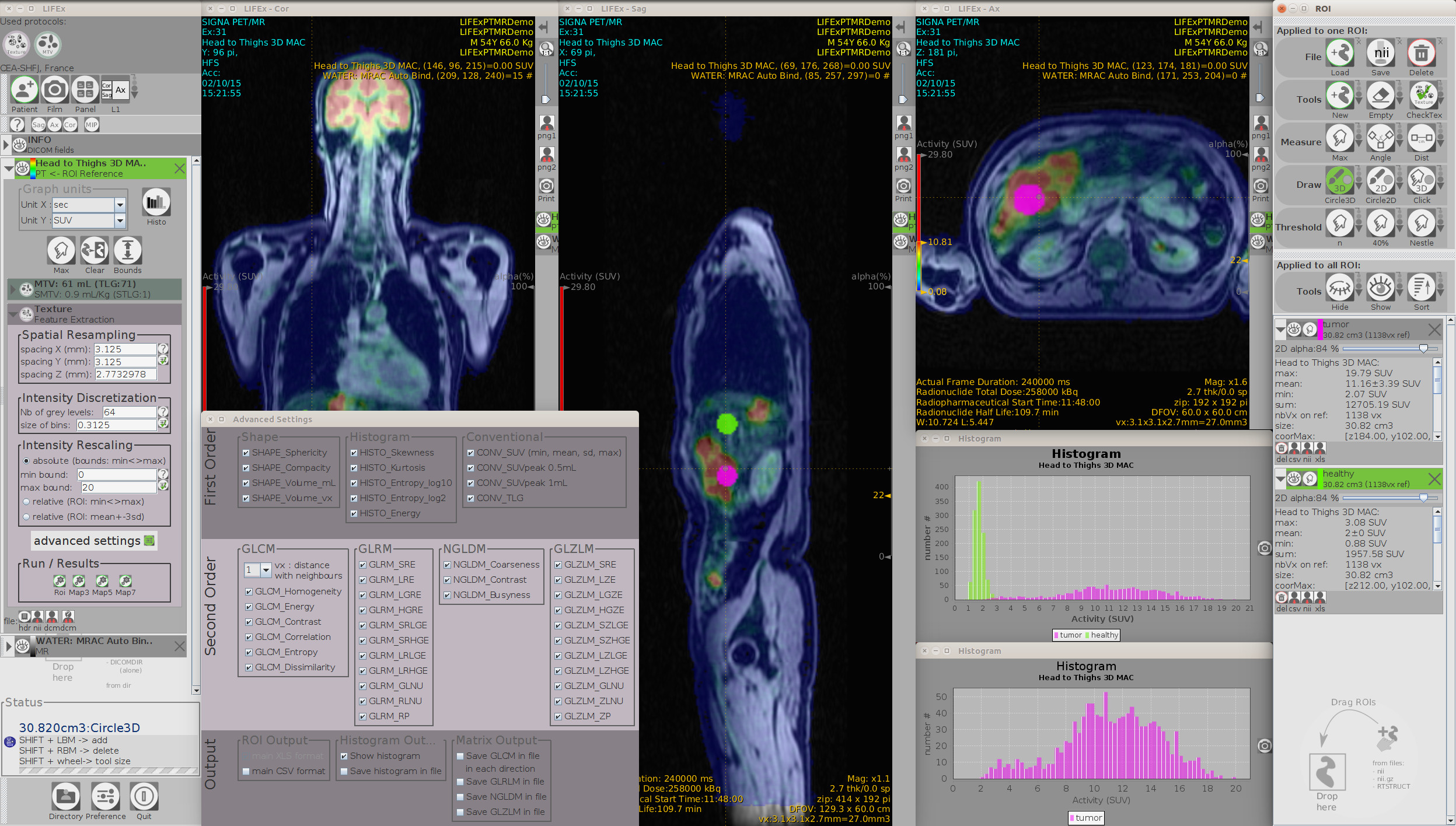
LIFEx v25.06.1 now available
LIFEx reads DICOM images locally or over a network using a DICOM browser and includes a powerful 3D reconstruction-based slice viewer. Volume of interest (VOI) can be either imported from previously created files or drawn and manipulated using LIFEx. Results are exported in Excel format files. LIFEx runs on Windows, MacOs and Linux. It is distributed with examples and includes a tutorial. User support is offered. Users can optionally contribute to the gathering of index values in different tissue types and different images as a public data bank of reference values is currently being built and integrated to the software for assisting the users with the interpretation of their results.
Download here : Download
LIFEx, Cancer Research 2018
LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity
C Nioche, F Orlhac, S Boughdad, S Reuzé, J Goya-Outi, C Robert, C Pellot-Barakat, M Soussan, F Frouin, and I Buvat. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Research 2018; 78(16):4786-4789
LIFExAnalytics
Location of user cities (monthly update):
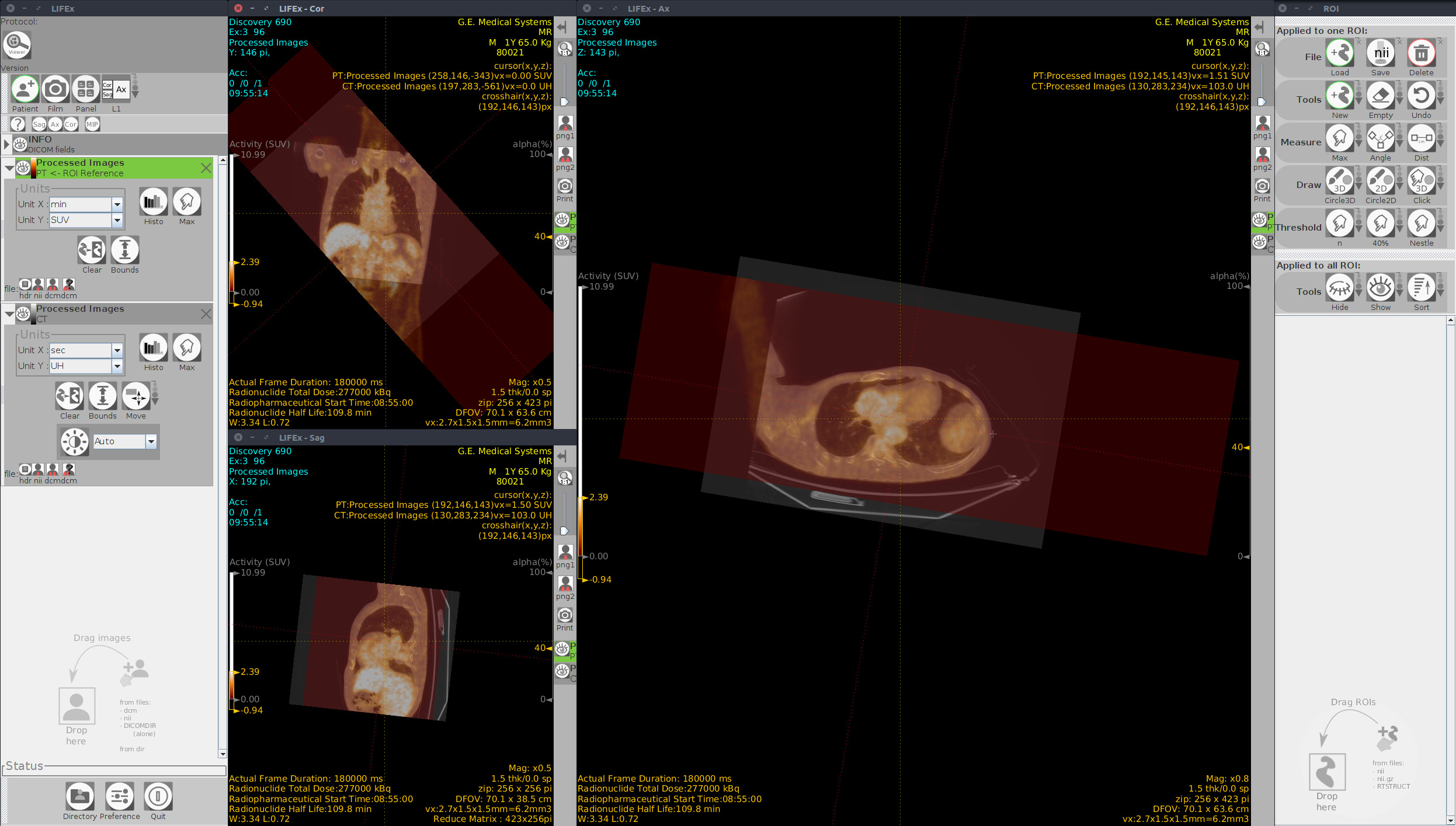
Screenshot MIP demos
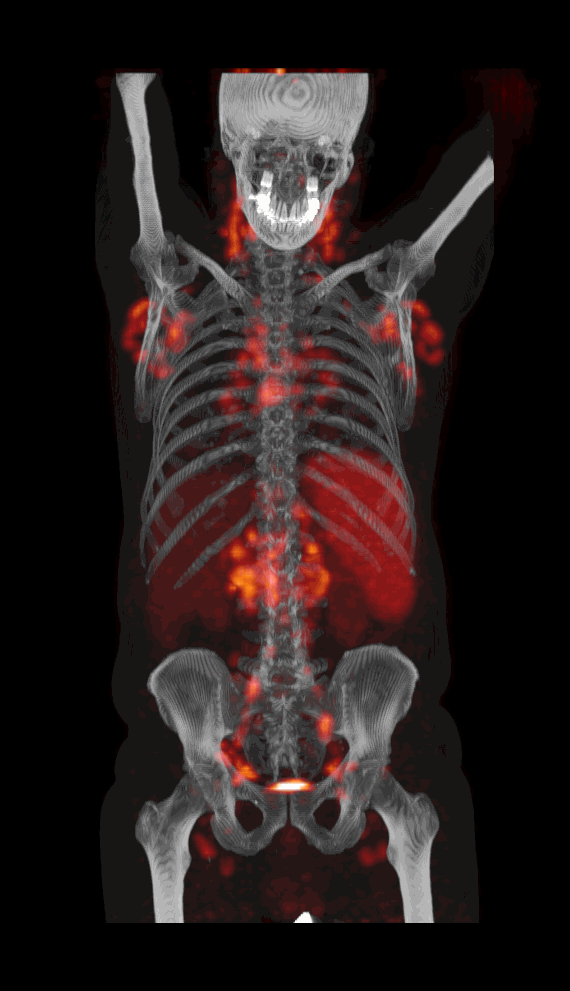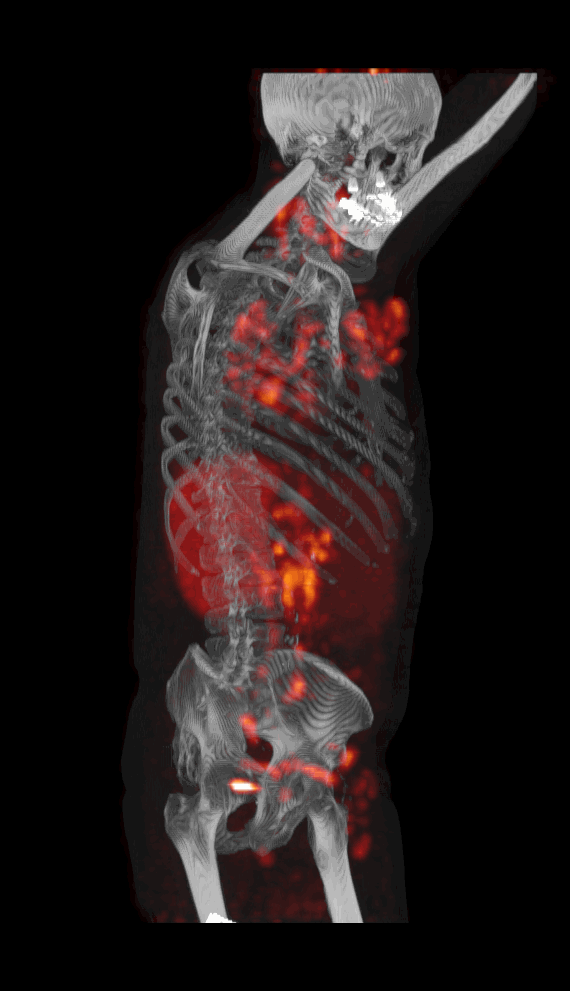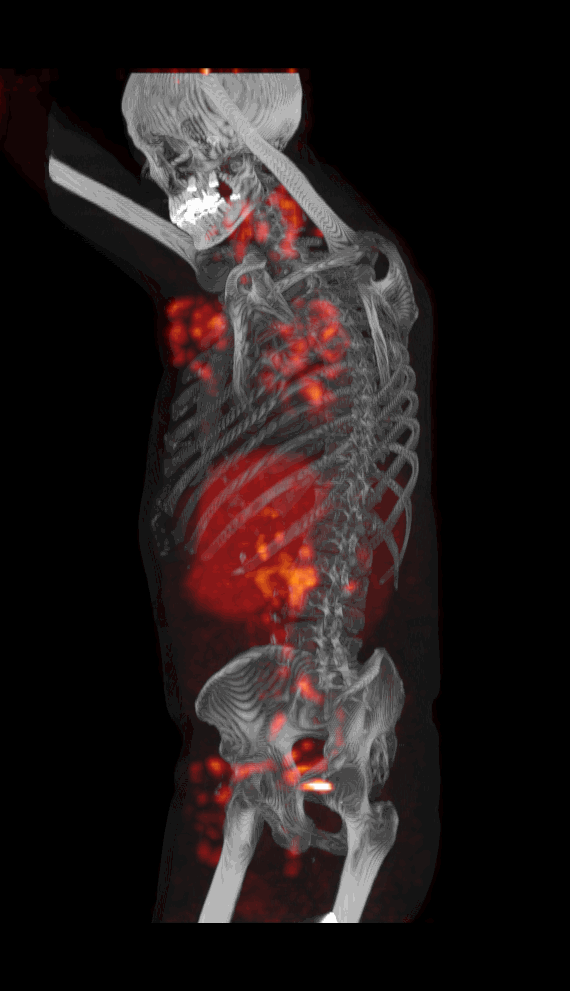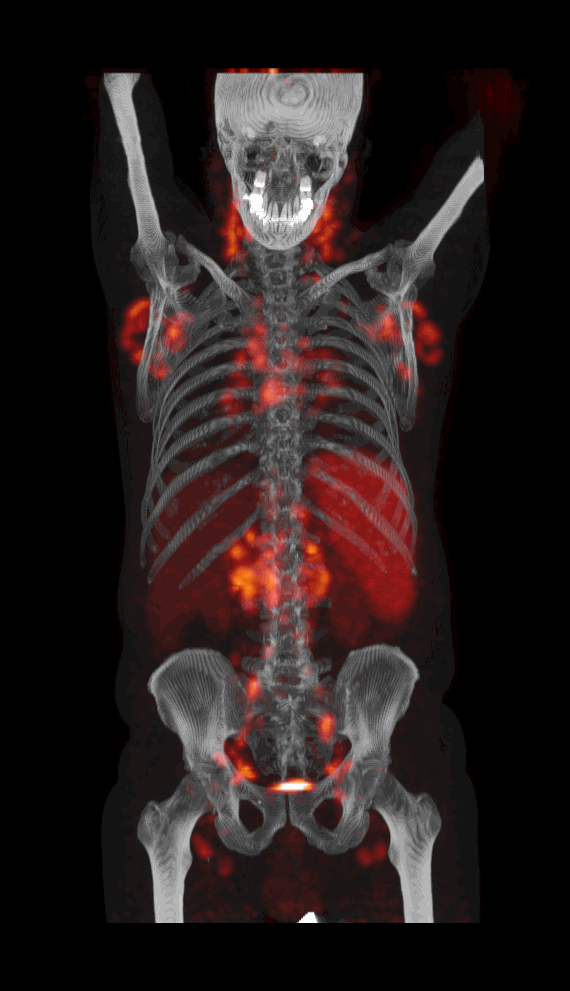 |
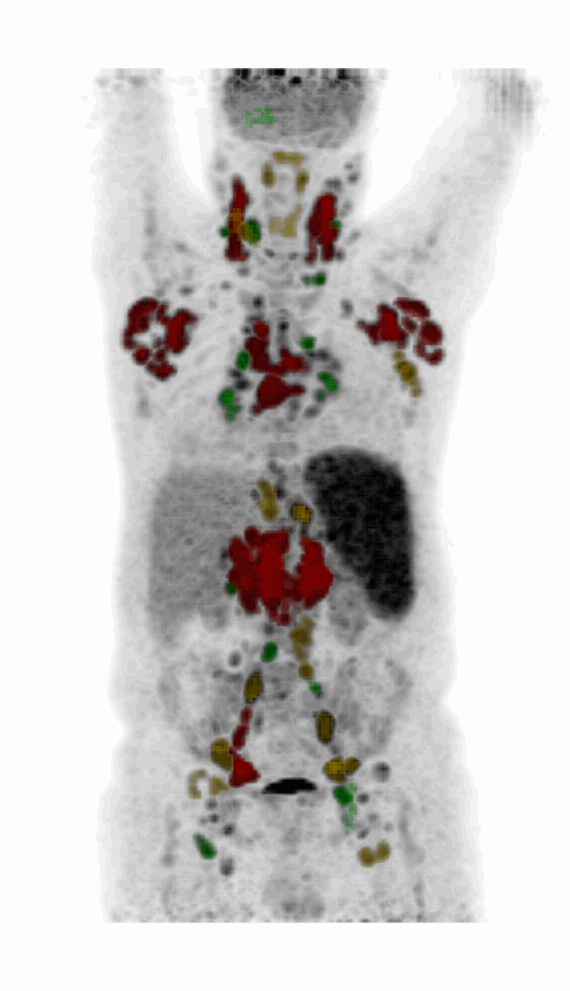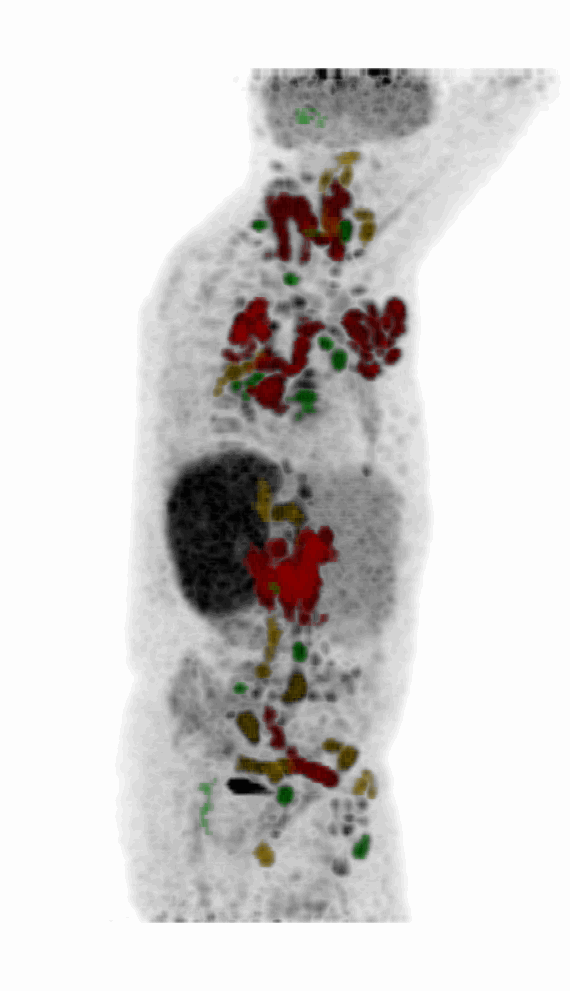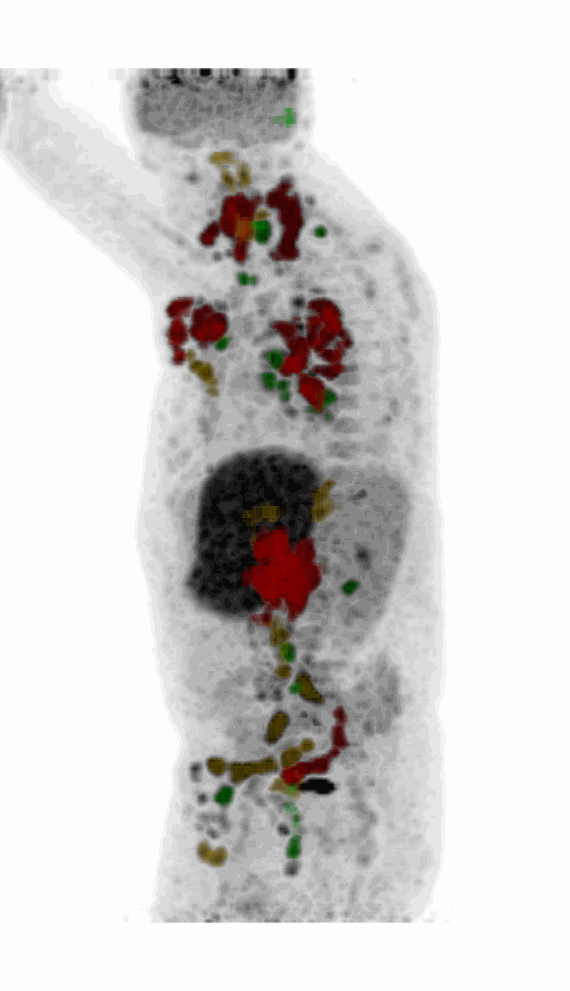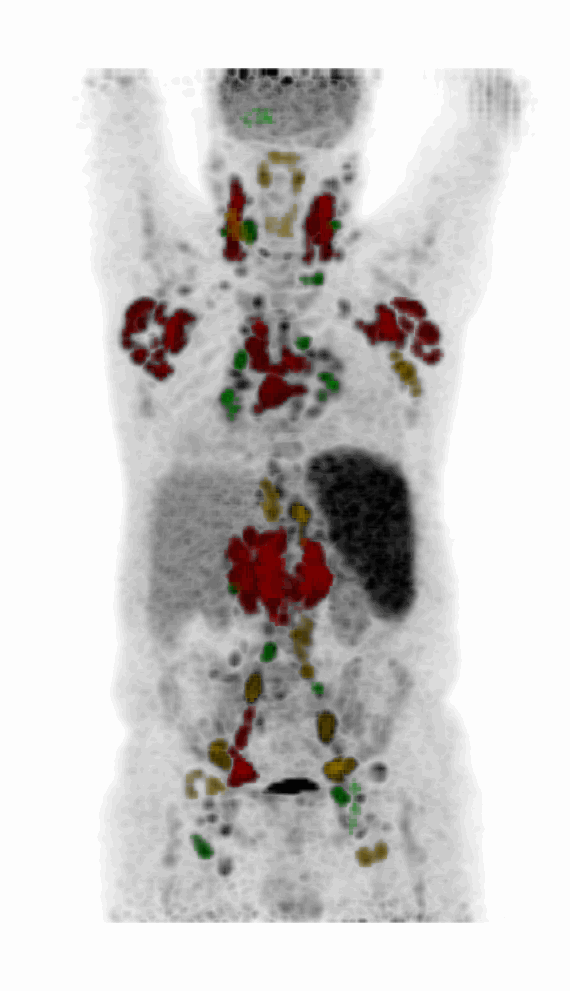 |
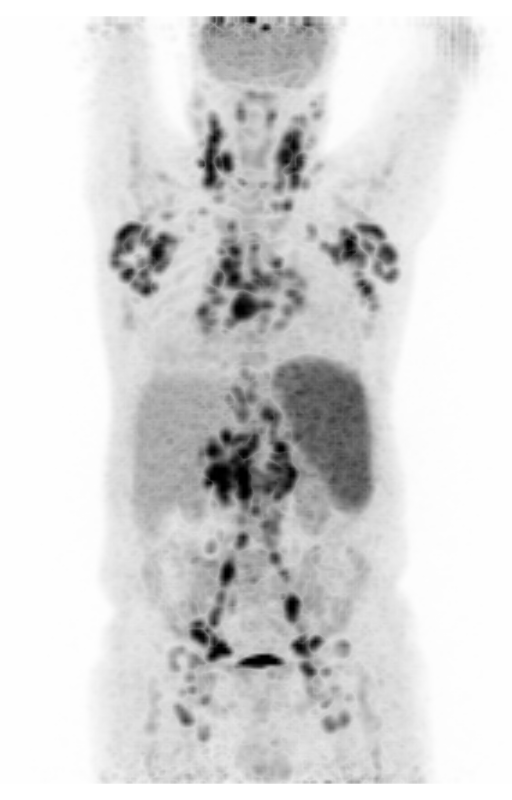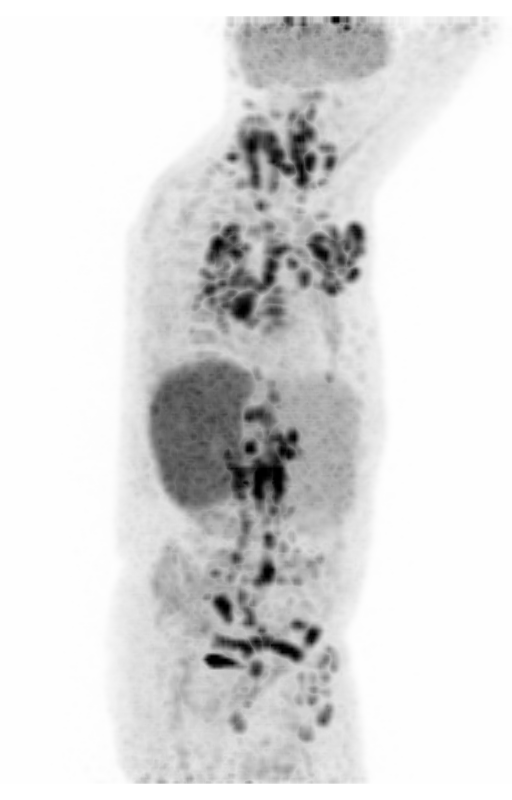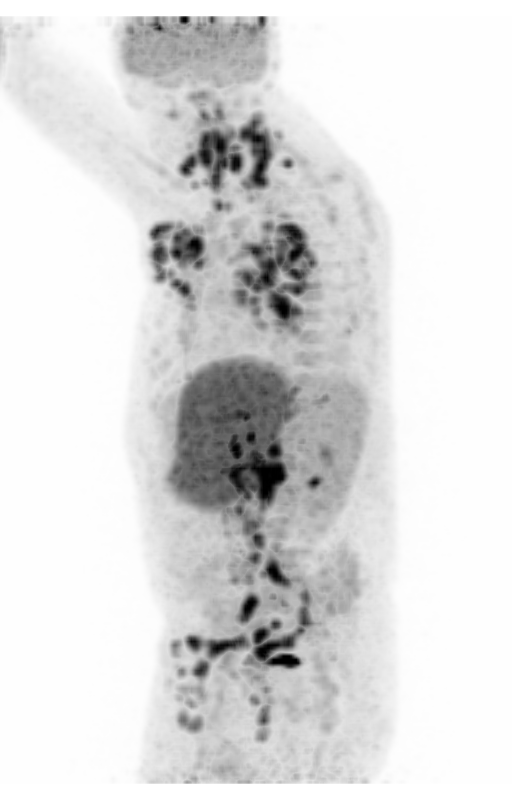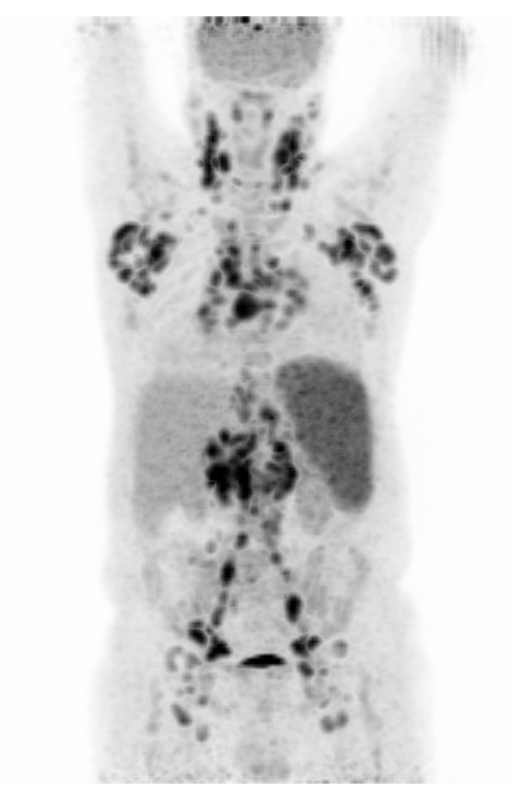 |
| MIP PET/CT | MIP PET/ROI | MIP PET |
Popular Features 2025
- reads nifti (.nii), ecat (.v), DICOM / RTSTRUCT (.dcm) files, and many others...
- creates 2D or 3D ROI
- analyses 311 histogram-based, image feature, mtv, summary and compare features
- complies with the Image biomarker standardisation initiative (IBSI)
- calculates total metabolic volume in PET
- MIP visualization
- writes results in Comma Separated Value (.csv)
- numerous publications (>1000 at 2025-09)
- large community of users (more than 10.000 users)
Meet us
You can always meet us at:
- 2026 SNMMI Annual Meeting, Los Angeles, USA from May 30 - June 2, 2026
- EANM'25, Annual Congress of the European Association of Nuclear Medicine, October 4 – 8, 2025, International Barcelona Convention Center (CCIB)
- at Institut Curie Centre De Recherche, Centre Universitaire, Bâtiment 101B, Rue Henri Becquerel, 91898 ORSAY CEDEX, 91400, Orsay, France
We are also attending many scientific meetings and conferences where you can meet us.
- ESTRO meeting 2026, 15 May 2026 - 19 May 2026 Stockholm, Sweden
If you want to meet us there, please send us an email at