Before asking questions or reporting problems to the contact address, please make sure your question has not been answered in the past.
Is LIFEx a free software?
Yes, it is free and all protocols are Open sources
How can I download it?
Please see the download page.
How to cite LIFEx?
Please see the How to cite LIFEx? page.
Which system for LIFEX run on?
The following OS are supported: MacOs (El Capitan, Maverick, Sierra, BigSur) (and for new M1 (aarch64) CPU of Mac), Windows (7, 8, 10), Linux (Fedora, RedHat, Ubuntu, Debian, Mint). No conflicts between system libraries and those of the binary packages exist.
I have installed an old version, can I reinstall over it without removing the previous version?
Yes you can. But if you run into problems, remove the old version(s) installed.
Wrong size windows, or windows out of place?
Convert a text file or .csv file into an Excel spreadsheet (.xls or .xslsx)
https://www.hesa.ac.uk/support/user-guides/import-csv
Please follow the instructions given at convert csv to columns
I can't find the button to save ROIs or Series any longer. Where is this button now?
Before (v<7.0.0):

For more homogeneity of the application, all actions are now called from the main menus (ROI menu for ROI, Series menu for series). The save button is now in the File/Edit of the submenu:
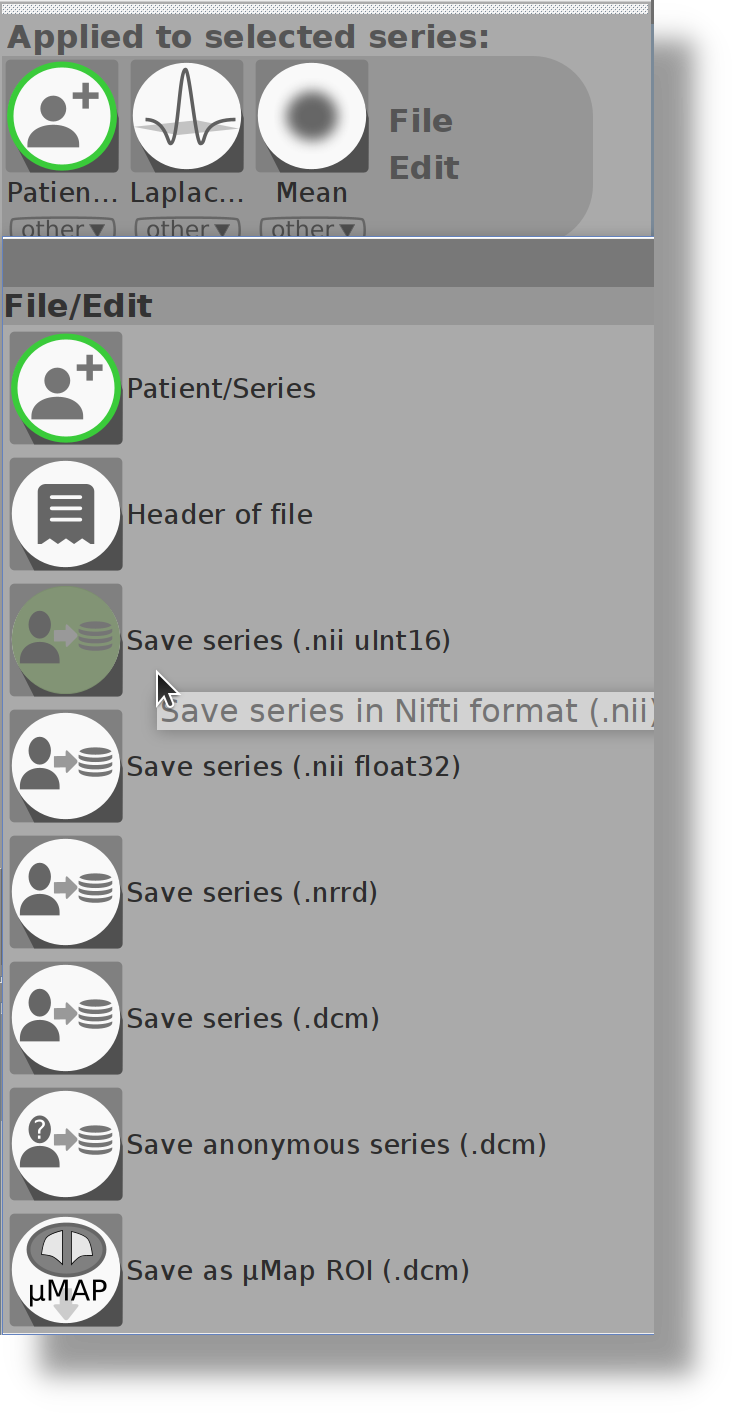
|
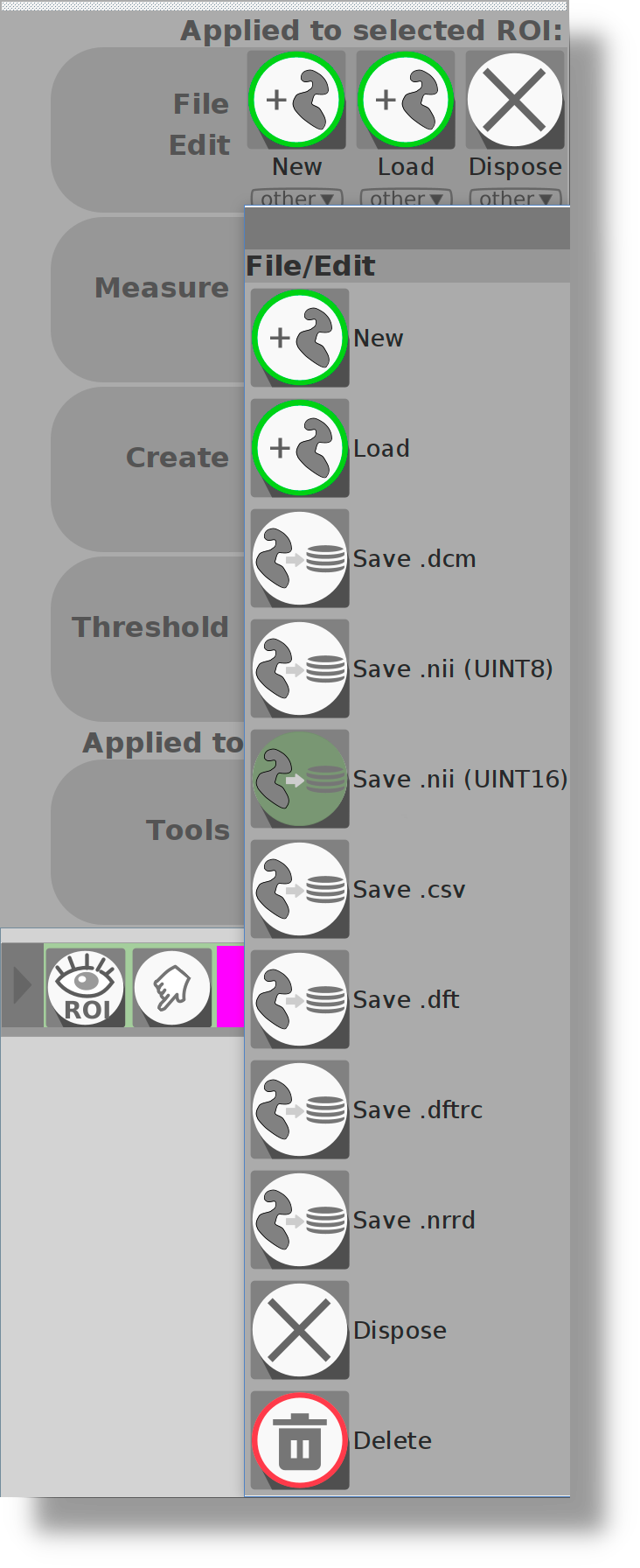
|
In PET, the DICOM fields that have to be properly filled for correct SUV correction are:
DICOM fields to SUV conversion :
(0008,0032) TM Acquisition Time
(0010,1030) DS Patient’s Weight
(0018,1074) DS Radionuclide Total Dose
(0018,1075) DS Radionuclide Half Life
(0018,1072) TM Radiopharmaceutical Start Time
(0054,1001) CS Units
(0028,1052) DS Rescale Intercept
(0028,1053) DS Rescale Slope
(0054,1102) DS DecayCorrection
I have been noticing that it has been taking a while to load my dicom images. Is there anything I can do to speed things up, or is there something off on my end?
There are several answers depending on the following cases:
- you don't have a lot of RAM on your computer (<=16 GB, often in laptops) then you need to quit all the applications that were open before LIFEx was opened! LIFEx will take what memory is left.
- you have too many applications at computer start-up that are already taking up all the memory (often on laptops, too). You need to remove these applications at start-up, as they are often not used enough (updates, etc.).
- you are trying to open images from an external USB disk: never.... copy the entire directory to your local computer disk before reading these images.
- you have a lot of RAM (>16 GB), so it's the DICOM images themselves that are causing LIFEx problems. In that case, we're interested in testing these images on my computer. Could you please send them to us.
Drag and drop does not work on Windows :
Change with regedit programme "EnableLUA=0" in HKEY_LOCAL_MACHINE/SOFTWARE/Microsoft/Windows/CurrentVersion/Policies/System/

